Abstract
Introduction
In the acute myeloid leukemia (AML) setting, artificial intelligence has mainly been used to facilitate diagnosis or to identify biological subcategories. In this work, we trained and compared machine learning and deep learning predictive models of outcome on the data of 3687 consecutive adult AML patients included in the DATAML registry between 2000 and 2019. We also trained a model to predict the best treatment for newly diagnosed AML over 70 years.
Methods
Feature engineering and selection were done to keep the most relevant variables among clinical and biological characteristics at diagnosis. We worked with 54 features per patient, as well as information about the treatment received (intensive chemotherapy (IC) or azacitidine (AZA)), response and survival.
We compared the performance of a gradient boosting algorithm (XGBoost) and three neural networks architectures: a multilayer perceptron (MLP), a neural oblivious decision ensemble model (NODE) and a recurrent relational network (RRN). We calibrated XGBoost with a grid search algorithm, and used 5-fold cross-validation on the dataset to evaluate all the models.
The Shapley Additive Explanations method (SHAP) was used to showcase the importance and influence of variables on the predictions. The Boruta algorithm was then used to extract the most important features for prediction.
Results
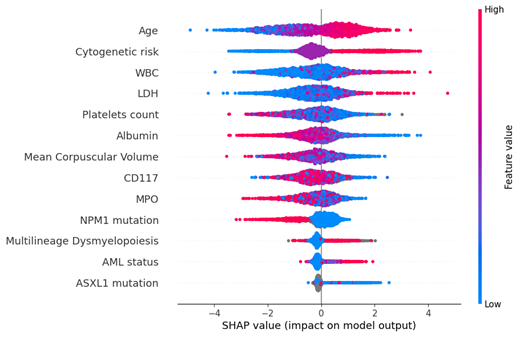
In our cohort, 3030 patients (82.2%) received IC and 657 (17.8%) AZA as first line treatment. Median overall survival (OS) was 18 and 9 months, respectively. We first designed models for OS prediction. In the IC cohort, we achieved an accuracy of 68.5% on predicting OS at the 18-month mark, an improvement of 17.5% over a naïve predictor. The Boruta algorithm selected 13 variables as the most important, with decreasing order of importance: age, cytogenetic risk, WBC, LDH, platelets count, albumin, MPO, mean corpuscular volume, CD117, NPM1 mutation, AML status, multilineage dysmyelopoiesis, ASXL1 mutation (Figure 1). When training with only these 13 variables, we achieved an accuracy of 67.8%.
In the AZA cohort, we achieved an accuracy of 62.1% on predicting OS at the 9-month mark, an improvement of 11.1% over a naïve predictor. Here the Boruta algorithm selected only 7 variables: blood blasts, serum ferritin, CD56, LDH, hemoglobin, CD13 and the presence of a disseminated intravascular coagulation. When training with only these 7 variables, we achieved a 61.9% accuracy.
We then designed models to predict the best treatment between IC and AZA for the 1032 patients older than 70 years. We achieved a 88.5% accuracy, which is 37.5% more than a naïve predictor given the distribution of the cohort: 51% having received IC and 49% having received AZA. For this model, 12 features out of 54 were selected by the Boruta algorithm as the most important: age, TP53 mutation, bone marrow blasts, AML status, disseminated intravascular coagulation, blood blasts, cytogenetic risk, IDH2 mutation, IDH1 mutation, presence of an infection at diagnosis, ASXL1 mutation and presence of leukostasis.
Conclusion
We show that predictive models can be trained on our database to predict with characteristics at diagnosis the treatment that would be chosen by an expert hematologist between IC and AZA in newly diagnosed AML, give an indication of OS with each treatment, and outperform classical statistical analysis or naïve predictors.
For the task of predicting OS, the improvement over naïve predictors is maximal at the median time of OS. We show with the Boruta algorithm that a small number of variables can recapitulate the accuracy of neural networks, which renders this type of model of high interest for routine practice, especially with the advent of targeted therapies.
Vergez: Pierre Fabre Laboratory: Research Funding; Roche: Research Funding. Dumas: BMS Celgene: Consultancy; Astellas: Consultancy; Daiichi-Sankyo: Consultancy. Tavitian: Novartis: Consultancy. Delabesse: Astellas: Consultancy; Novartis: Consultancy. Pigneux: Amgen: Consultancy; Sunesis: Consultancy, Research Funding; BMS Celgene: Consultancy, Research Funding; Roche: Consultancy, Research Funding; Novartis: Consultancy, Research Funding. Recher: Pfizer: Honoraria, Membership on an entity's Board of Directors or advisory committees; Novartis: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Macrogenics: Honoraria, Membership on an entity's Board of Directors or advisory committees; MaatPharma: Research Funding; Jazz: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Janssen: Honoraria; Incyte: Honoraria; Daiichi Sankyo: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; BMS/Celgene: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Astellas: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Amgen: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Roche: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Takeda: Honoraria, Membership on an entity's Board of Directors or advisory committees; Agios: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; AbbVie: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding. Bertoli: Astellas: Consultancy; BMS Celgene: Consultancy; Abbvie: Consultancy; Jazz Pharmaceuticals: Consultancy.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal